Abstract
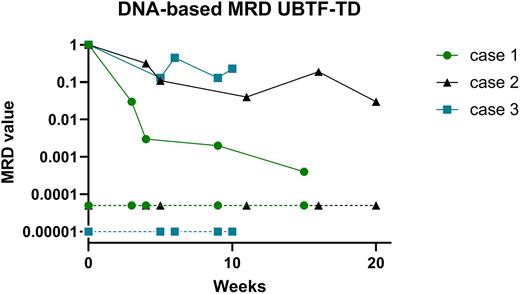
Pediatric acute myeloid (AML) is considered as a heterogeneous disease affecting 15% of children with acute leukemia. Outcome is still unsatisfactory with an overall survival around 70-80% and a relapse rate of 30-40% after primary treatment. The most important factors determining clinical outcome are genetic aberrations, and early response to treatment. Many studies have demonstrated the prognostic significance of minimal residual disease (MRD) monitoring. Although MRD monitoring is now an integral part in the management (including MRD-guided stratification) of Acute Lymphoblastic Leukemia (ALL) in children (generally done by ASO-qPCR on tumor specific IG/TCR targets), its clinical utility in childhood non-lymphoblastic leukemia (such as AML) is currently unknown/not used. Contemporary approaches to MRD detection in AML rely on PCR amplification of usually fixed genetic abnormalities (MRD-PCR) or on flowcytometric profiling of leukemia-associated immunophenotypes (LAIP). Molecular MRD detection is in general more sensitive than flow-based MRD, but the applicability is lower since recurrent mutational targets (e.g. fusion genes) using conventional technology, are present in only 40-50% of pediatric AML patients. This RNA-based MRD is common practice for CBFb-MYH11, RUNX1-RUNXT1, KMT2A-AFF1, -MLLT1, -MLLT3, and NPM1 exon 12 mutations and standard curves necessary for MRD quantification are available for all mentioned genes. Since the introduction of Whole Transcriptome Sequencing (WTS; RNAseq) as standard diagnostics in our laboratory, we identified various additional fusion genes in AML (increasing available molecular targets up to 75%), but also in other non-B cell malignancies such as SCLL, CEL, HES and BPDCN. By designing patient specific primers/probe sets surrounding the breakpoint sequences, we were able to generate 36 RNA-based MRD assays (including FOP-FGFR1, RANBP2-ALK, DEK-NUP214, ETV6-PDGFRB, KMT2A-AFDN) and monitor MRD during/after therapy. Although RNA-based MRD approach allows additional molecular monitoring in these patients, this type of MRD displays several disadvantages: sensitivity of a certain MRD assay can be low due to low expression levels of the fusion gene or low RNA yield and/or quality. Furthermore, quantification can be hampered by lack of available standard curves. Therefore DNA-based MRD is favorable over RNA-based MRD and Whole Exome Sequencing (WES) or Whole Genomic Sequencing (WGS) can identify the genomic breakpoint sequence in respectively genomic exons or introns. In addition, other genomic aberrations (such as insertions, deletions and point mutations) can be identified by these techniques and used for development of DNA-based MRD. By using this approach we were able to generate 17 additional DNA-based MRD assays (including FOP-FGFR1, RANBP2-ALK, RUNX1-CBFA2T3 NPM1-MLF1 UBTF-TD, p53 deletion), all with very high sensitivity (e.g. around 10-4 to 5x10-5) and patient-derived standard curves to quantify MRD. Figure 1 shows an example for DNA-based MRD monitoring of 3 AML patients harboring the recently described poor-outcome UBTF-TD aberration showing no/slow decrease of MRD in all patients. In conclusion, we show that fusion gene detection by WTS, preferably in combination with mutation analysis by WES/WGS, is providing comprehensive diagnostic results to improve the development of highly sensitive patient specific MRD targets to allow better molecular monitoring (and prediction of relapse), eventually leading to treatment stratification of patients in the future. We will present an update of our results and interesting cases at the upcoming conference.
Figure 1: By WES, 3 patients with tandem duplications (TD) in exon 13 of the UBTF gene were identified, and patient specific primers and probes were designed for MRD on follow-up DNA samples. MRD was quantified by using a standard curve derived from serial dilutions from the diagnostic DNA. Quantitative range, sensitivity and background were determined according to the ESG-MRD-ALL guidelines.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal